Postdoctoral fellows are a crucial part of Van Andel Institute’s efforts to improve the health and enhance the lives of current and future generations.
All fellows conduct their work in the Institute’s state-of-the-art research space and have access to an extensive suite of Core Technologies and Services as well as a dedicated team of operations and sponsored research professionals. They also have the opportunity to network with colleagues from around the world, who come to VAI as part of the many invited lectures, scientific symposia and other events held throughout the year.
The Institute recognizes that selecting where to complete a postdoctoral fellowship is a significant decision for postdoctoral fellows and their families. Located in Grand Rapids, Michigan, VAI is committed to providing postdoctoral fellows with a high-caliber experience that prepares them for successful futures as scientific leaders. VAI’s dedicated Office of Postdoctoral Affairs provides a comprehensive professional development program. In addition, the Institute also provides a competitive benefits package for all postdoctoral fellows, regardless of funding source.
Learn more about VAI’s postdoctoral program
Questions? Email [email protected]. Please note, the Institute does not currently award external fellowships.
Research
Openings
Interested in becoming a postdoc at VAI? Visit our Career Center to view current openings or contact one of our faculty members directly.


Research
Postdoctoral Fellows
Working on an individual personalized project under the guidance of a qualified faculty mentor, VAI’s exceptional postdoctoral fellows make great contributions to the science conducted in the Institute’s laboratories. They play a crucial role in the intellectual development and management of research projects and contribute to a considerable share of the hands-on research activities in the laboratory.
Research
Living in Grand Rapids
Whether you’re in search of a beach, a craft brew, unique cuisine or the world’s biggest art competition, Grand Rapids and West Michigan have something for everyone.

Testimonials
As a registered Patent Agent with the U.S. Patent and Trademark Office, I work in diverse fields of science and technology in pursuit of protecting our clients’ intellectual property. As a postdoctoral fellow, VAI’s multi-disciplinary approach to research allowed me to continue to hone my analytical and technical communication skills, which are invaluable in the practice of intellectual property protection.
Jessica Hessler, Ph.D.
Patent Agent, Price HeneveldAs a postdoc at VAI, the skills I gained in grant writing and review, working collaboratively, as well as experience with cutting-edge resources has helped me in managing peer-review committees and providing scientific oversight of research grants. The intimate setting at VAI offered opportunities for building unique relationships, which were invaluable in navigating my career path.
Heather Calderone, Ph.D.
Science Manager, American Brain Tumor AssociationWhen I think about my experiences as a VAI postdoc, I am most grateful for the support and flexibility that allowed me to explore and gain experience as an educator in higher education. Without this opportunity, I would not have considered a career in medical education. The opportunity to teach and conduct research while a postdoc helped me feel supported beyond the laboratory, and has led to a rewarding career as an educator.
Schoen W. Kruse, PhD, F-NAOME
Associate Dean of Integrated Learning, College of Osteopathic Medicine, Associate Professor of Pharmacology, Kansas City University of Medicine and BiosciencesAs a VAI postdoctoral fellow in Dr. Jeff MacKeigan’s Laboratory of Systems Biology, I was able to acquire and hone critical skills in advanced techniques, project development, grant writing and laboratory management. VAI’s postdoctoral program fostered an environment of productive creativity and, together with VAI’s facilities, resources and support, this made my time there thoroughly enjoyable and successful.
Nathan Lanning, Ph.D.
Assistant Professor, Department of Biological Sciences, California State University, Los AngelesIt was a wonderful experience to work at VAI. I had a postdoctoral fellowship in Dr. Michael Weinreich’s Laboratory of Chromosome Replication (2005-2008). It was a very good experience because I could acquire a lot of skills and advanced techniques at the professional level but also maturity at the personal level. This opportunity opened me doors when I returned in France because this mobility to VAI was very valuable to find a permanent position.

Dorine Bonte, Ph.D.
Assistant Professor, College of Pharmacy, Paris-Sud University, FranceI remember the spirit of collaboration and innovation at Van Andel Institute. That environment enabled me to partner with fellow VAI scientists to learn and apply new (at the time) genomics techniques to my scientific questions. The experience of genomics discovery opened my eyes to new scientific possibilities and led to a career at a leading genomics company, where I now share similar stories of genomics discoveries with my podcast listeners.
Paul Bromann, Ph.D.
Staff Scientific Liaison and Host of the Illumina Genomics Podcast, Scientific Affairs, Illumina, Inc.My postdoctoral fellowship was mentored by Dr. Nick Duesbery, back in 2000-2001; these first years of the Institute were bathed in an incomparable atmosphere of creativity and productivity. Interacting with researchers from different fields opened my mind. This fellowship enabled me to grow up in autonomy; while I learnt new technical skills, I was allowed to acquire and discern the abilities and values associated with being at the helm of a scientific team.
Prof. Jean-François Bodart
Vice-Dean for International Relations, Professor, Faculty of Sciences and Technologies, University of LilleI acquired so many important skills during my time as a VAI postdoc. In addition to developing additional research and technical skills in the lab, I was provided training in grant writing, lab management and communication, among others. All of these now help me immensely in my current role. I would have been walking into this position blindly if it weren’t for the excellent training and programming provided by the VAI OPA! Jennifer Lamberts, Ph.D. Assistant Professor, College of Pharmacy, Ferris State University
Jennifer Lamberts, Ph.D.
Assistant Professor, College of Pharmacy, Ferris State UniversityI would describe my time as a postdoc at VAI as very exciting and full of opportunities. There was support and funding in place for postdocs to invite speakers, organize events and gain teaching experience, which are essential for career development.
Aikseng Ooi, Ph.D.
Assistant Professor, College of Pharmacy, University of ArizonaVan Andel Institute (VAI) offers several types of postdoctoral positions. Each is designed to prepare early career scientists for the next steps in their careers and offers competitive funding and benefits.
Postdoctoral Fellows
VAI is strongly committed to providing postdoctoral fellows with a high-caliber experience that prepares them for successful futures as scientific leaders. Through mentorship by one or more of the Institute’s accomplished faculty, postdoctoral fellows gain the skills necessary to launch independent careers.
In addition to the Institute’s extensive scientific resources, postdoctoral fellows have access to a dedicated support team of professionals who can aid in everything from fellowship applications to experimental design. The Office of Postdoctoral Affairs also provides a comprehensive professional development program. Learn more about VAI postdoctoral fellows’ demographic and career outcomes here.
Interested in a postdoctoral fellowship at VAI? We are always accepting applications. Please contact the investigator directly for more information on opportunities to join the team. Specific openings are available in our Careers section.
Most postdocs at VAI are full-time employees at the Institute and, as such, receive a competitive salary and benefits package, including health insurance and 401(k) opportunities.
VAI encourages, but does not require, all postdocs to apply for external fellowship funding. We provide numerous resources to assist with the application process and an incentive award for those who receive awards. In addition, VAI offers internal funding opportunities through our T32-funded Cancer Epigenetics Training (CET) program and our Inspire Fellowship.
Salary
VAI has a minimum salary schedule that begins at $70,000 and increases annually based on satisfactory performance. Pay runs from $70,000 to $95,000 for those with specialized skill sets or above-average qualifications. Pay begins at $95,000 for those with highly specialized skill sets (e.g., bioinformatics).
Benefits
VAI offers a comprehensive set of benefits including:
- Medical, dental and vision plan choices for self and family at a minimal cost
- Institute-paid life and accidental death and dismemberment insurance
- Voluntary term life and accidental death and dismemberment insurance for self and family
- Institute-paid short-term and long-term disability insurance (includes maternity leave)
- Flexible spending accounts for dependent care and medical expenses
- Holiday pay
- Vacation days and paid time off for illness and personal needs
- Six weeks of paid parental leave
- Long-term care insurance
- Tuition reimbursement
- Opportunity to participate in a 401(k) plan
- Incentive award for achieving external grant/fellowship funding
- Professional development
- Employee Assistance Services
In addition to a maximum reimbursement of $3,000 for moving expenses, postdoctoral fellows have access to VAI’s contracted relocation coordinator, who can assist with relocation needs such as housing, establishing a bank account and applying for a social security card.
In addition to VAI’s top-of-the-line research facilities, the Institute also offers:
- An on-site café that provides affordable breakfast and lunch options
- A 24/7 fitness center with a variety of workout equipment
- A library with on-site publications as well as access to books and papers housed at other academic institutions
- Dedicated parking
VAI Fellows
As part of its commitment to develop the next generation of biomedical research leaders, the Institute is proud to support exceptional early career scientists through its VAI Fellows program, which offers highly competitive, enhanced fellowships with a greater degree of freedom than traditional postdoctoral training opportunities.
VAI Fellows receive additional financial and research support coupled with increased independence, which allows them to singularly focus on their own novel, interdisciplinary, high-impact projects. They also benefit from having multiple faculty mentors who can provide guidance to ensure success, both in the lab and in the business of science.
VAI Fellows slots are highly competitive. Potential applicants should reach out to the principal investigator of their choice and indicate interest in the program. Applications are reviewed by the Faculty Advisory Committee to the Chief Scientific Officer. Applicants who do not receive a VAI Fellowship may still be offered a different postdoctoral fellow position. A list of VAI’s labs and investigators may be found here.
VAI Fellows receive a greater degree of autonomy to develop their own independent research programs, supported by mentorship from VAI’s expert faculty and a separate research budget.
Most postdocs at VAI are full-time employees at the Institute and, as such, receive a competitive salary and benefits package, including health insurance and 401(k) opportunities.
Salary and Research Funding
VAI Fellows receive a greater degree of autonomy to develop their own independent research programs, supported by mentorship from VAI’s expert faculty, an enhanced salary and a separate research budget. Figures are based on the applicant’s experience.
Benefits
VAI offers a comprehensive set of benefits including:
- Medical, dental and vision plan choices for self and family at a minimal cost
- Institute-paid life and accidental death and dismemberment insurance
- Voluntary term life and accidental death and dismemberment insurance for self and family
- Institute-paid short-term and long-term disability insurance (includes maternity leave)
- Flexible spending accounts for dependent care and medical expenses
- Holiday pay
- Vacation days and paid time off for illness and personal needs
- Six weeks of paid parental leave
- Long-term care insurance
- Tuition reimbursement
- Opportunity to participate in a 401(k) plan
- Professional development
- Employee Assistance Services
In addition to a maximum reimbursement of $3,000 for moving expenses, postdoctoral fellows have access to VAI’s contracted relocation coordinator, who can assist with relocation needs such as housing, establishing a bank account and applying for a social security card.
In addition to VAI’s top-of-the-line research facilities, the Institute also offers:
- An on-site café that provides affordable breakfast and lunch options
- A 24/7 fitness center with a variety of workout equipment
- A library with on-site publications as well as access to books and papers housed at other academic institutions
- Dedicated parking
Postdoctoral Research Fellows
Some postdoctoral fellowship sponsors do not allow VAI to employ the recipient postdoc. Instead, VAI appoints those funded by these awards as postdoctoral research fellows and provides them with a competitive stipend and benefits package.
VAI is strongly committed to providing postdoctoral research fellows with a high caliber experience that prepares them for successful futures as scientific leaders. Through mentorship by one or more of the Institute’s accomplished faculty, postdoctoral research fellows gain the skills necessary to launch independent careers.
In addition to the Institute’s extensive scientific resources, postdoctoral research fellows have access to a dedicated support team of professionals who can aid in everything from fellowship applications to experimental design. The Office of Postdoctoral Affairs also provides a comprehensive professional development program. Learn more about VAI postdoctoral fellows’ demographic and career outcomes here.
Postdoctoral Research Fellows receive a competitive stipend and benefits package, including health insurance.
Stipend
VAI has a minimum salary schedule that begins at $70,000 and increases annually based on satisfactory performance. Pay runs from $70,000 to $95,000 for those with specialized skill sets or above-average qualifications. Pay begins at $95,000 for those with highly specialized skill sets (e.g., bioinformatics).
Benefits
VAI offers a comprehensive set of benefits including:
- Medical, dental and vision plan choices for self and family at a minimal cost
- Institute-paid life and accidental death and dismemberment insurance
- Short-term disability coverage (includes maternity leave)
- Holiday pay
- Vacation days and paid time off for illness and personal needs
- At least six weeks of paid parental leave
- Stipend supplementation plus an incentive award for achieving external grant/fellowship funding
- Professional development
- Employee Assistance Services
In addition to a maximum reimbursement of $3,000 for moving expenses, postdoctoral research fellows have access to VAI’s contracted relocation coordinator, who can assist with relocation needs such as housing, establishing a bank account and applying for a social security card.
In addition to VAI’s top-of-the-line research facilities, the Institute also offers:
- An on-site café that provides affordable breakfast and lunch options
- A 24/7 fitness center with a variety of workout equipment
- A library with on-site publications as well as access to books and papers housed at other academic institutions
- Dedicated parking
Looking for world-class postdoctoral training in cancer epigenetics? You’ve come to the right place.
Van Andel Institute is an internationally recognized center of excellence for basic and translational epigenetics research. The Institute’s Cancer Epigenetics Training (CET) Program prepares postdoctoral fellows to become independent research leaders in the field of cancer epigenetics. The program offers cutting-edge, interdisciplinary research training in structural and functional aspects of epigenetics, complemented by state-of-the-art Core Technologies and Services, unique professional development activities, and integration with established translational research programs and partnerships led by VAI investigators. Trainees will emerge from the program with comprehensive training in cancer epigenetics and a professional foundation from which to launch their independent research careers.
As part of the program, trainees will be able to pursue basic and translational research projects in:
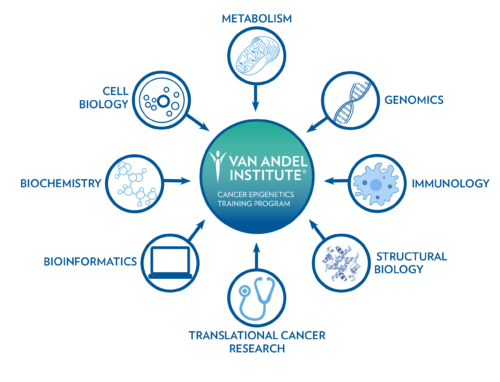
- Cancer biology
- Tumorigenesis
- Cellular development and differentiation
- Mechanisms governing DNA methylation and chromatin regulation
- Epigenomics and advanced tool development
- GWAS and EWAS
- Transgenerational inheritance
- Structural biology
- Metabolism
- Epigenetic therapies
Trainees also will have exclusive opportunities to participate in the Van Andel Institute–Stand Up To Cancer Epigenetics Dream Team, a multi-institutional effort to move promising combination therapies for cancer into clinical trials.
Questions? Please contact Dr. Erica Gobrogge, program director of VAI’s Office of Postdoctoral Affairs, at [email protected].
The Cancer Epigenetics Training program is supported by a National Cancer Institute T32 training grant (no. T32CA251066). The content is solely the responsibility of VAI and does not necessarily represent the official views of the National Institutes of Health.
VAI’s Cancer Epigenetics Training Program accepts applications on a rolling basis. Start your application by clicking “Apply Now” above.
Eligibility: Applicants must be near completion of a Ph.D., M.D. or other appropriate terminal degree. Applicants must be U.S. citizens or permanent residents. Individuals who identify as underrepresented minorities are strongly encouraged to apply.
Prior to beginning the application, please review the list of Cancer Epigenetics Training Program faculty members (found under the “Faculty” tab) to determine whose research interests you, and prepare the following required materials:
- NIH-style biosketch
- Research statement: The statement should comprise a high-level description of your proposed research project, the significance of the problem and its potential impact on the field. It should follow the format of an NIH-style specific aims page (you can find guidance here).
- Three reference letters (one should be from your thesis advisor or a thesis committee member)
If you have already spoken with a VAI faculty member about joining their lab or applied for one of our open postdoc positions, there will be space to indicate this on the application. This is encouraged but not required to apply to the Cancer Epigenetics Training Program.
Please note, you may reuse these materials to apply for our Postdoc Preview (learn more here) and our Inspire Program, for those who identify as underrepresented minorities (learn more here).
VAI’s Cancer Epigenetics Training Program is an interdisciplinary postdoctoral training program that provides trainees with extensive research and mentorship opportunities in the field of cancer epigenetics.
Upon acceptance, each trainee is assigned an Individual Development Team (IDT) that will coach, mentor and guide them through the scientific and non-scientific dimensions of career development. The IDT comprises the trainee’s primary mentor, a secondary mentor and another faculty member (either a non-CET Program VAI faculty member or a non-VAI faculty member). Please see the list of mentors below.
Throughout the program, trainees will complete a comprehensive curriculum of scientific and professional development training. Trainees will be expected to:
- Present annual seminars (internal and external)
- Complete at least one semester of teaching or mentoring experience
- Publish at least one first-authored publication
- Submit F32 (or equivalent) and K99/R00 applications
Peter A. Jones, Ph.D., D.Sc. (hon)
Cancer Epigenetics Training Program Director
Professor and Chief Scientific Officer
Scott Rothbart, Ph.D.
Cancer Epigenetics Training Program Associate Director
Professor, Department of Epigenetics
Yvonne Fondufe-Mittendorf, Ph.D.
Professor, Department of Epigenetics
Stephanie Grainger, Ph.D.
Assistant Professor, Department of Cell Biology
Russell Jones, Ph.D.
Chair and Professor, Department of Metabolism and Nutritional Programming
Connie Krawczyk, Ph.D.
Associate Professor, Department of Metabolism and Nutritional Programming
Peter Laird, Ph.D.
Professor, Department of Epigenetics
Huilin Li, Ph.D.
Chair and Professor, Department of Structural Biology
Gerd Pfeifer, Ph.D.
Professor, Department of Epigenetics
Andrew Pospisilik, Ph.D.
Professor, Department of Epigenetics
Hui Shen, Ph.D.
Professor, Department of Epigenetics
Xiaobing Shi, Ph.D.
Chair and Professor, Department of Epigenetics
Timothy Triche, Jr., Ph.D.
Associate Professor, Department of Epigenetics
Hong Wen, Ph.D.
Professor, Department of Epigenetics
Evan Worden, Ph.D.
Assistant Professor, Department of Structural Biology
Co-mentors
Stephen Baylin, M.D.
Director’s Scholar
Adelheid (Heidi) Lempradl, Ph.D.
Assistant Professor, Department of Metabolism and Nutritional Programming
Evan Lien, Ph.D.
Assistant Professor, Department of Metabolism and Nutritional Programming
Questions? Contact Dr. Erica Gobrogge, program director of VAI’s Office of Postdoctoral Affairs and a CET leadership team member, at [email protected].
Do I need to submit a separate employee application along with my Cancer Epigenetics Training Program application?
Applicants who currently do not have a postdoctoral position at VAI should first submit the Cancer Epigenetics Training Program application. Details on required materials may be found under the “Apply” tab. The application may be found here.
We encourage you to apply to any open postdoctoral positions of interest and/or contact mentors of interest directly via email after you submit your CET application. You may find open positions on VAI’s Career Center.
How many applicants are accepted each year?
The Cancer Epigenetics Training Program will accept two new trainees each year.
Who reviews applications? When will I find out if I’m accepted?
Applications will be reviewed by the Cancer Epigenetics Training Program leadership team and program faculty members. We will notify all applicants regarding the outcome of their application. Please be patient since applications are accepted on a rolling basis.
I’m also interested in VAI’s Postdoc Preview and/or the Inspire Fellowship. How do I apply?
Applications for Postdoc Preview, the Cancer Epigenetics Training Program and the Inspire Program are separate. However, applicants may reuse the same materials for each application (for example, you may reuse a Cancer Epigenetics Training Program reference letter for the other applications). Please double-check the application requirements for each program, as they may have additional required components.
How is the CET program different than a traditional VAI postdoc?
CET trainees build an Individual Development Team, have exclusive opportunities to participate in the VAI-Stand Up to Cancer Epigenetics Dream Team, complete a comprehensive, formal scientific and professional development training curriculum, and receive a unique stipend and benefits package.
Helpful Links
VAI Postdoctoral Fellowships home
Mary Reyland, Ph.D. (Advisory Committee Chair)
Professor, University of Colorado Anschutz Medical Campus
Jerry Callahan, Ph.D., MBA
Chief Strategic Officer, Van Andel Institute
James Fahner, M.D.
Founding and current Division Chief of Pediatric Hematology/Oncology, Helen DeVos Children’s Hospital
Associate Professor, Department of Pediatrics/Human Development, Michigan State University
Kay Macleod, Ph.D.
Associate Professor, University of Chicago
Danny Welch, Ph.D.
Professor and Associate Director for Cancer Education and Career Enhancement, University of Kansas Cancer Center
Supporting the next generation of research leaders through rigorous, personalized postdoctoral training
Van Andel Institute’s Inspire Fellowship supports exceptional postdoctoral fellows by reducing barriers, fostering community, and providing the resources and personalized mentorship needed for an outstanding training experience. We are committed to ensuring fellows have a firm foundation from which to launch their independent research careers — and to make a world of difference. Applications for the Inspire Fellowship are accepted year round and reviewed on a rolling basis.
Inspire Fellowships offer:
- Funding that totals $150,000 over 2-3 years
- Cutting-edge research training
- One-on-one mentorship from a team of VAI faculty
- Individualized professional development
- A community of peers
- Access to outstanding Core Technologies and Services
Questions? Please contact Dr. Erica Gobrogge, program director of VAI’s Office of Postdoctoral Affairs, at [email protected].
Eligibility: Applicants must be within 12 months of completing a Ph.D., M.D. or other appropriate terminal degree. Applicants must be U.S. citizens or permanent residents who identify as members of an underrepresented racial and/or ethnic group as defined by the National Institutes of Health.
Prior to beginning the application, please prepare the following required materials:
- Curriculum vitae or NIH-style biosketch.
- Personal statement: Please provide an overview of your scientific background, your goals for your postdoctoral training and why you are interested in VAI and the Inspire Fellowship program. You also may include any personal information you feel is relevant to the evaluation of your application. The statement should be no more than three pages.
- One reference letter
- A list of VAI faculty members with whom you are interested in working. If you have already spoken with a VAI faculty member about joining their lab or have applied for one of our open postdoc positions, there will be space to indicate this on your application. Corresponding with a VAI faculty member prior to applying for the Inspire Fellowship is encouraged but not required.
Applications for VAI postdoctoral positions and the Inspire Fellowship are separate. Receipt of the Inspire Fellowship is contingent on an offer of employment for a postdoctoral position in a VAI lab. We encourage Inspire applicants to apply to open postdoctoral positions of interest and/or contact faculty mentors of interest directly via email after submitting the Inspire Fellowship application.
Please note: You may reuse these materials to apply for our Cancer Epigenetics Training Program (learn more here).
Do I need to submit a separate employment application along with my Inspire Fellowship application?
Applicants who currently do not have a postdoctoral position at VAI should first submit the Inspire Fellowship application. Details on required materials may be found under the “Apply” tab. The application may be found here.
We encourage you to apply to any open postdoctoral positions of interest and/or contact faculty mentors of interest directly via email after you submit your Inspire Fellowship application. You may find open positions on VAI’s Career Center.
Who reviews applications? When will I find out if I’m accepted?
Applications will be reviewed by the Inspire Fellowship steering committee, comprised of VAI faculty and Office of Postdoctoral Affairs leadership.
I’m also interested in VAI’s Postdoc Preview and/or the Cancer Epigenetics Training Program Fellowship. How do I apply?
Applications for Postdoc Preview, the Cancer Epigenetics Training Program and the Inspire Fellowship are separate. However, applicants may reuse the same materials for each application (for example, you may reuse an Inspire Fellowship reference letter for the other applications). Please double-check the application requirements for each program, as they may have additional required components.
See the requirements for the Postdoc Preview here and for the Cancer Epigenetics Training Program here.
Van Andel Institute is dedicated to excellence in research and scientific training.
As a reflection of this commitment, we are pleased to join the Coalition for Next Generation Life Sciences, a collaborative effort to enhance transparency by publishing outcomes data for graduate students and postdoctoral fellows.
Metrics include
- Admissions and matriculation data of Ph.D. students
- Median time-to-degree and completion data for Ph.D. programs
- Demographics of Ph.D. students and postdoctoral scholars by gender, underrepresented minority status and citizenship status
- Median time in postdoctoral status at the institution
- Career outcomes for Ph.D. and postdoctoral alumni, classified by job sector and career type using a common taxonomy. Source: Coalition for Next Generation Life Sciences
Learn more about the Coalition by visiting nglscoalition.org.
For questions about VAI graduate student data, please contact Christy Mayo at [email protected]. For questions about VAI postdoctoral fellow data, please contact Dr. Erica Gobrogge at [email protected].
Graduate Student Statistics
Current Graduate Students
Numbers current as of Sept. 1, 2023
By Gender
Data represents current student population
Total students: 51
Race/Ethnicity for USC/PR
Data does not include international students
Total students: 32
Citizenship
Data represents current student population
Total students: 51
Admissions
Fall 2023 Cohort
Applicant Pool
Time to Degree(TTD)
By Year of Graduation (in Three Year Aggregates)
By Demographic Categories
Retention, Attrition & Complete Outcomes
Outcomes in Multi-Year Aggregates of Entering Cohorts
Please hover over the chart to see the data.
Aggregate Outcomes by Gender*
Please hover over the chart to see the data.
*Through 2017 entering cohort, as of August 2023
Ph.D. Career Outcomes by Career Type*
Please hover over the chart to see the data.
*Through 2017 entering cohort, as of August 2023
Ph.D. Career Outcomes by Job Sector*
Please hover over the chart to see the data.
*Through 2017 entering cohort, as of August 2023
Postdoctoral Fellow Statistics
Please hover over each chart to see data.
Van Andel Institute Definition of a Postdoctoral Fellow/Fellowship
The fellowship is a limited time after receipt of a doctoral degree for mentored and advanced training through research under the supervision of a principal investigator (mentor). The postdoctoral fellow, in mutual agreement with their mentor, undertakes an individual research project and service, teaching and professional development activities that together will provide the experience and knowledge essential for career advancement and establishing an independent career.
By Gender
As of Jan. 1 of each year.
Citizenship Status
As of Jan. 1 of each year.
Race/Ethnicity for USC/PR
As of Jan. 1 of each year. Data does not include international postdocs.
Time as a Postdoctoral Fellow
Median Time in Postdoc Training by Gender
Median Time in Postdoc Training by Nationality
Postdoctoral Fellows’ Career Outcomes Based on Last Known Position
Sector
Includes all VAI posdocs (n=221) who have completed training as of Feb. 28, 2024 (UCOT v2017).
Career Type
Includes all VAI posdocs (n=221) who have completed training as of Feb. 28, 2024 (UCOT v2017).
Job Function
Includes all VAI posdocs (n=221) who have completed training as of Feb. 28, 2024 (UCOT v2017).
Are you a senior graduate student exploring your postdoc options?
VAI Postdoc Preview provides selected applicants the opportunity to visit Van Andel Institute to learn about our postdoctoral training opportunities, meet one-on-one with our internationally recognized faculty and explore our extensive scientific resources, including our state-of-the-art Core Technologies and Services.
VAI offers two Postdoc Preview events each year—one in the spring and one in the fall. Our Spring application deadline has passed. We will post more information about the Fall event in early summer. There is no cost to attend for selected applicants; VAI provides transportation, lodging and meals. We also are pleased to offer family care grants for those caring for children or other family members. More information is below.
About postdoctoral training at VAI
VAI labs conduct world-class biomedical research in epigenetics, neurodegeneration, metabolism, cell biology and structural biology — all supported by outstanding Core Technologies and Services. Postdocs are a crucial part of VAI, so they receive highly competitive salaries, which start at $70,000 per year. VAI also offers postdocs comprehensive benefits, including low-cost health insurance for self and family, paid parental leave, and much more.
We offer numerous fully funded postdoctoral positions across our areas of focus. Learn more about postdoctoral training opportunities at VAI here. In addition, we have two internal fellowship programs: a National Cancer Institute T32-funded Cancer Epigenetics Training (CET) program, and the Inspire Fellowship program for postdocs who identify as a member of underrepresented groups. Please note that attending Postdoc Preview does not guarantee you a fellowship or a postdoc position at VAI; we encourage you to also apply to one or more of our open postdoc positions and fellowship opportunities (see FAQ tab below for more information).
Questions? Please contact Dr. Erica Gobrogge, program director of VAI’s Office of Postdoctoral Affairs, at [email protected].
Eligibility
Applicants must be current graduate students at U.S. institutions who anticipate beginning a postdoctoral fellowship within the next year. Individuals who identify as members of underrepresented groups are strongly encouraged to apply. Preference will be given to applicants who are eligible for VAI’s NCI T32-funded Cancer Epigenetics Training Program and VAI’s Inspire Fellowship.
How to Apply
VAI offers two Postdoc Preview events each year—one in the spring and one in the fall. Our Spring application deadline has passed. We will post more information about the Fall event in early summer. Prior to starting the application, please compile a list of VAI faculty members whose research interests you, along with your curriculum vitae.
What can you expect during a Postdoc Preview visit?
Day 1
6:00–8:00 p.m. | Welcome dinner with VAI postdocs
Day 2
9:00–10:00 a.m. | Welcome presentation and breakfast
10:00–11:00 a.m. | Five-minute flash talks from participants
11:00 a.m. – 12:00 p.m. | One-on-one meetings with faculty
12:00–1:00 p.m. | Lunch with current postdocs
1:00–2:00 p.m. | Tour of VAI
2:00–3:30 p.m. | One-on-one meetings with faculty
3:30–5:00 p.m. | VAI Research-in-Progress seminar and Refresh
6:30–8:30 p.m. | Dinner with faculty
Day 3
Explore Grand Rapids and return home
Do I need to apply to attend?
Yes. Postdoc Preview selects attendees through an application process. See the Apply section to learn more.
Is there a cost to attend?
There is no cost to attend for selected applicants! VAI will arrange your transportation, lodging and meals and will reimburse you for reasonable additional expenses you incur (e.g., airport parking).
I would like to attend but I have child or family care needs. Should I still apply?
Yes! VAI is pleased to offer family care grants of up to $500 to selected applicants. For more information, please contact Eric Miller at [email protected].
If selected, do I get to choose the faculty with whom I meet?
Yes! As part of your application, we ask that you select faculty members you would like to meet. We will do our best to pair you with the faculty of your choosing, depending on their availability. You can learn more about our faculty by visiting VAI’s lab directory or by exploring each department’s home page:
- Department of Epigenetics
- Department of Neurodegenerative Science
- Department of Cell Biology
- Department of Metabolism and Nutritional Programming
- Department of Structural Biology
If I am selected to attend the Postdoc Preview, am I guaranteed a postdoctoral position at VAI?
No. Postdoc Preview offers an opportunity to learn about postdoctoral training opportunities at VAI, meet potential faculty mentors and network with other future postdocs, all while exploring Grand Rapids. We encourage you to apply to our different postdoc programs either before or after you attend Postdoc Preview. You can learn more about our postdoctoral offerings and how to submit an application here.
Who reviews applications?
Applications will be reviewed by VAI faculty members and VAI Office of Postdoctoral Affairs staff. We are particularly interested in applicants whose research interests fit with our faculty members’ research.
Meet the Postdocs
Working on an individual personalized project under the guidance of a qualified faculty mentor, VAI’s exceptional postdoctoral fellows make great contributions to the science conducted in the Institute’s laboratories. They play a crucial role in the intellectual development and management of research projects and contribute to a considerable share of the hands-on research activities in the laboratory.
Department of Epigenetics

Reid Blanchett, Ph.D.


Sophia Chaudhry, Ph.D.

Brooke Grimaldi, Ph.D.

Tim Gruber, Ph.D.

Youssef Hegazy, Ph.D.

Joel Hrit, Ph.D.
Regulatory mechanisms of maintenance DNA methylation by DNMT1

Josh (Hyo Sik) Jang, Ph.D.
Single cell resolution of immune landscape in cancer therapy


Junwei Niu, Ph.D.

Smitha George, Ph.D.

Elisson Lopes, Ph.D.

Nour El Osmani, Ph.D.
Xiaoyan Xie, Ph.D.
The role of DNA methylation in controlling gene expression during cancer development
DEPARTMENT OF NEURODEGENERATIVE SCIENCE

Manish, Ph.D.

Ehraz Anis, Ph.D.
Exploring the contribution of the vermiform appendix to Parkinson’s disease


Clarissa Ferolla, Ph.D.


Vera Martin Masegosa, Ph.D.

Caio Massari, Ph.D.

Alice Prigent, Ph.D.

Jordan Rowlands, Ph.D.

Dorian Sargent, Ph.D.
Exploring the mechanisms implicating VPS35 in the neuropathology of Parkinson’s disease

Alex Soto-Avellaneda, Ph.D.


Carlos Vivaldo, Ph.D.

Saima Zameer, Ph.D, M.Pharm, B.Pharm
DEPARTMENT OF CELL BIOLOGY
DEPARTMENT OF STRUCTURAL BIOLOGY


Jinhong Hu, Ph.D.
Fei Jin, Ph.D.

Yangyang Li, Ph.D.

Anjali Malik, Ph.D.

Quoc Phong Nguyen, Ph.D.

Merissa (Xiansha) Xiao, Ph.D.
Proteasome and protein folding system involved in multi-drug resistance in Mycobacteria

Yuan Zeng, Ph.D.
DEPARTMENT OF METABOLISM AND NUTRITIONAL PROGRAMMING

Alix Booms, Ph.D.
Thesis: Uncovering the functionality of PD risk-SNPs at the HLA locus in microglia

Michael Dahabieh, Ph.D.
Characterizing heterogeneity of exhausted T cells and understanding the roles of peroxisomes in the tumor microenvironment

Rachel (Rae) House, Ph.D.
Thesis: Identification and exploitation of metabolic reprogramming in NF1-mutant breast cancer

Yujin Jeong, Ph.D.

Hyo-Jun Kim, Ph.D.

Marco Mechan Llontop, Ph.D.

Jacob Longenecker, Ph.D.


Kelsie Nauta, Ph.D.

Hemendra Panwar, Ph.D.

Eduardo Perez-Mojica, Ph.D.
Mechanistic dissection of epigenetic inheritance

Samuel Preston, Ph.D.

Krittika Sudhakar, Ph.D.
Thesis: The effect of maternal nutrition on offspring development and metabolism


Lukai Zhai, Ph.D.
Histone H3 lysine 4 demethylase KDM5C/D functions in dendritic cells



